- Gene activity in the brain is responsible for whether a species is monogamous
- New study covers 450 million years of evolution—a greater span than ever before
- Across multiple species, the same genes are turned on or off to cause monogamy
Why are some animals committed to their mates and others are not?
 According to a new study led by researchers at that looked at 10 species of vertebrates, evolution used a kind of universal formula for turning non-monogamous species into monogamous species — turning up the activity of some genes and turning down others in the brain.
According to a new study led by researchers at that looked at 10 species of vertebrates, evolution used a kind of universal formula for turning non-monogamous species into monogamous species — turning up the activity of some genes and turning down others in the brain.
"Our study spans 450 million years of evolution, which is how long ago all these species shared a common ancestor," said , research associate in UT Austin's and first author of the study published in the journal .
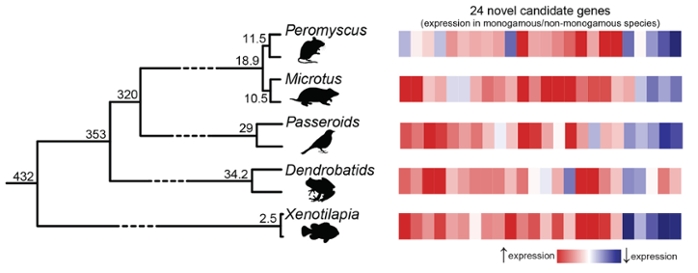
The authors define monogamy in animals as forming a pair bond with one mate for at least one mating season, sharing at least some of the work of raising offspring and defending young together from predators and other hazards. Researchers still consider animals monogamous if they occasionally mate with another.
The researchers studied five pairs of closely related species – four mammals, two birds, two frogs and two fish — each with one monogamous and one non-monogamous member. These five pairs represent five times in the evolution of vertebrates that monogamy independently arose, such as when the non-monogamous meadow voles and their close relatives the monogamous prairie voles diverged into two separate species.
The researchers compared gene expression in male brains of all 10 species to determine what changes occurred in each of the evolutionary transitions linked to the closely related animals. Despite the complexity of monogamy as a behavior, they found that the same changes in gene expression occurred each time. The finding suggests a level of order in how complex social behaviors come about through the way that genes are expressed in the brain.
This study covers a broader span of evolutionary time than had been explored previously. Other studies have looked at genetic differences related to evolutionary transitions to new traits, but they typically focus on animals separated by, at most, tens of millions of years of evolution, as opposed to the hundreds of millions of years examined with this study.
 "Most people wouldn't expect that across 450 million years, transitions to such complex behaviors would happen the same way every time," Young said.
"Most people wouldn't expect that across 450 million years, transitions to such complex behaviors would happen the same way every time," Young said.
Researchers examined gene activity across the genomes of the 10 species, using RNA-sequencing technology and tissue samples from three individuals of each species. The scientists detected gene-activity patterns across species using bioinformatics software and the .
Using a software package, , the team was able to arrange genes from distantly related species — such as a fish and a mammal — into groups based on sequence similarities. This allowed them to identify the common evolutionary formula that led to pair bonds and co-parenting in the five species that behave monogamously.
"Wrangler is set up with a relational database that allows individual computational steps to go back and talk to this database, and pull up the information it needs without any timeout errors," Young said. "We've been able to run all of our species together on Wrangler using OrthoMCL, and at this point we haven't even maxed out what Wrangler is capable of doing."
According to Young, with traditional online databases she was only able to identify about 350 comparable genes across these 10 species; however, when she ran OrthoMCL on Wrangler, she identified almost 2,000 genes that are comparable across all of the species.
"This is an enormous improvement from what is available," Young said. "When you add this up across 10 species, you have an enormous amount of data. We're starting at a minimum of 80,000 genes that we're going to compare in all pairwise combinations for over three billion comparisons to perform and organize in total. The staff at TACC and the Wrangler supercomputer helped make this science possible."
Read more:
